Wheat Variety Mixture Study: Automating Disease Detection through Machine Learning
For more details, please refer to the preprint submitted for peer review attached herewith.
Introduction
This document presents a comprehensive study on automating disease detection in wheat variety mixtures using machine learning. The project was conducted in collaboration with Clément Plessis, Axel Prouvost, and Léna Connesson, focusing on optimizing wheat variety mixtures to combat fungal diseases and improve yields in sustainable farming practices.
Summary
- 🌾 Overview
- 🖼️ Data Preprocessing
- 🤖 Machine Learning Model Development
- 📊 Model Evaluation
- 📈 Results
- 🚀 Real-world Application
- ✅ Conclusion
- 🚧 Challenges and Learnings
🌾 Overview
This project was conducted in collaboration with Clément Plessis as part of his PhD research on “Genomics and Evolution of Plant-Plant Interactions: Application to the Cultivation of Varietal Mixtures for Pesticide-Free Agriculture.” The focus of the study is on optimizing wheat variety mixtures, both with and without fungicides, to combat fungal diseases like yellow rust and oidium, while improving yields in sustainable farming practices.
Clément established 304 micro-plots, each containing a mixture of 8 wheat genotypes selected from 100 varieties. Additionally, 182 nursery plots were set up with a single genotype. These experiments were conducted under both fungicide-treated and untreated conditions, where a total of 8,000 wheat leaves were collected and scanned to assess infected areas and explore genotype interactions.
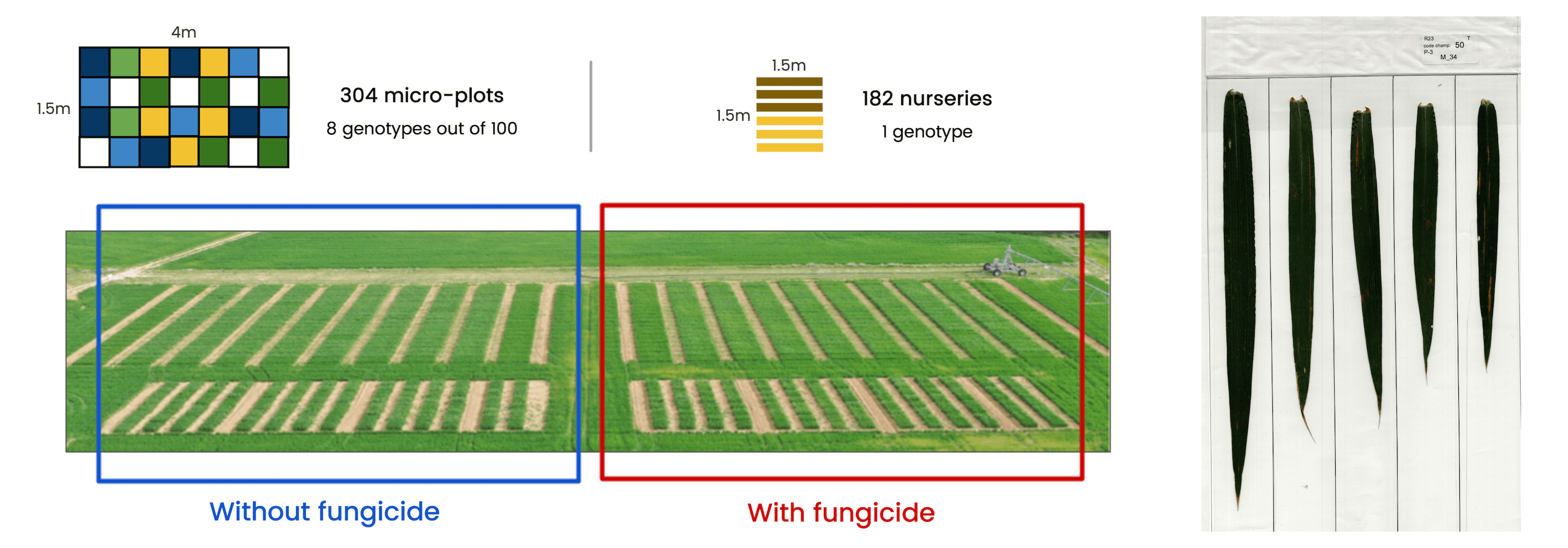
To enhance the analysis efficiency, we developed a machine learning solution capable of automating the analysis of these leaf samples. The manual evaluation process was labor-intensive and repetitive, so our goal was to streamline this workflow through automation.
🖼️ Data Preprocessing
The dataset consists of images with 5 wheat leaves per page, necessitating leaf extraction from each image before analysis. We used a series of image processing techniques to build the following pipeline:
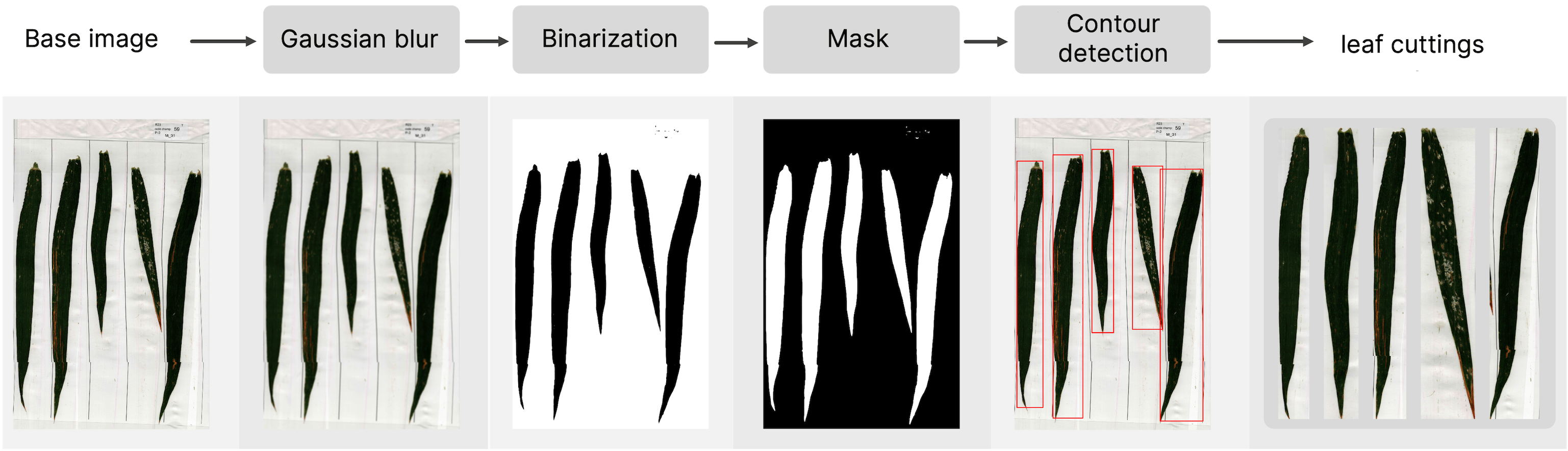
Each leaf was labeled with the respective plot and mixture number. To extract this information, we leveraged EasyOCR, a Python package based on the Tesseract engine, to perform Optical Character Recognition (OCR) on the labels present in the scanned images. This allowed us to automatically tag the leaves with relevant metadata.

🤖 Machine Learning Model Development
The aim of this project was to develop a machine learning model that could be run on everyday laptops without requiring powerful hardware or GPUs. Moreover, it was important that the model be retrainable by researchers with minimal machine learning expertise.
We chose Ilastik, a software tool that allows for easy pixel classification through manual annotation of images. However, Ilastik’s output was not readily compatible with Python workflows, so we developed a Python package called EasIlastik (2k downloads) to facilitate seamless integration of the Ilastik model into Python scripts.
Once trained, the model can be applied with just a few lines of code:
import EasIlastik
EasIlastik.run_ilastik(input_path = "path/to/input/folder",
model_path = "path/to/model.ilp",
result_base_path = "path/to/output/folder/")
📊 Model Evaluation
To evaluate the performance of our model, we used two distinct methods:
- Artificial Patches: We created synthetic patches with known infected and healthy areas to precisely control the extent of infection and accurately evaluate the model’s prediction accuracy.
- Ground Truth Dataset: We manually annotated 50 wheat leaves using VGG Image Annotator (VIA), creating a ground truth dataset. The model was then used to predict infected areas, and we compared the model’s results with the manually labeled ground truth.
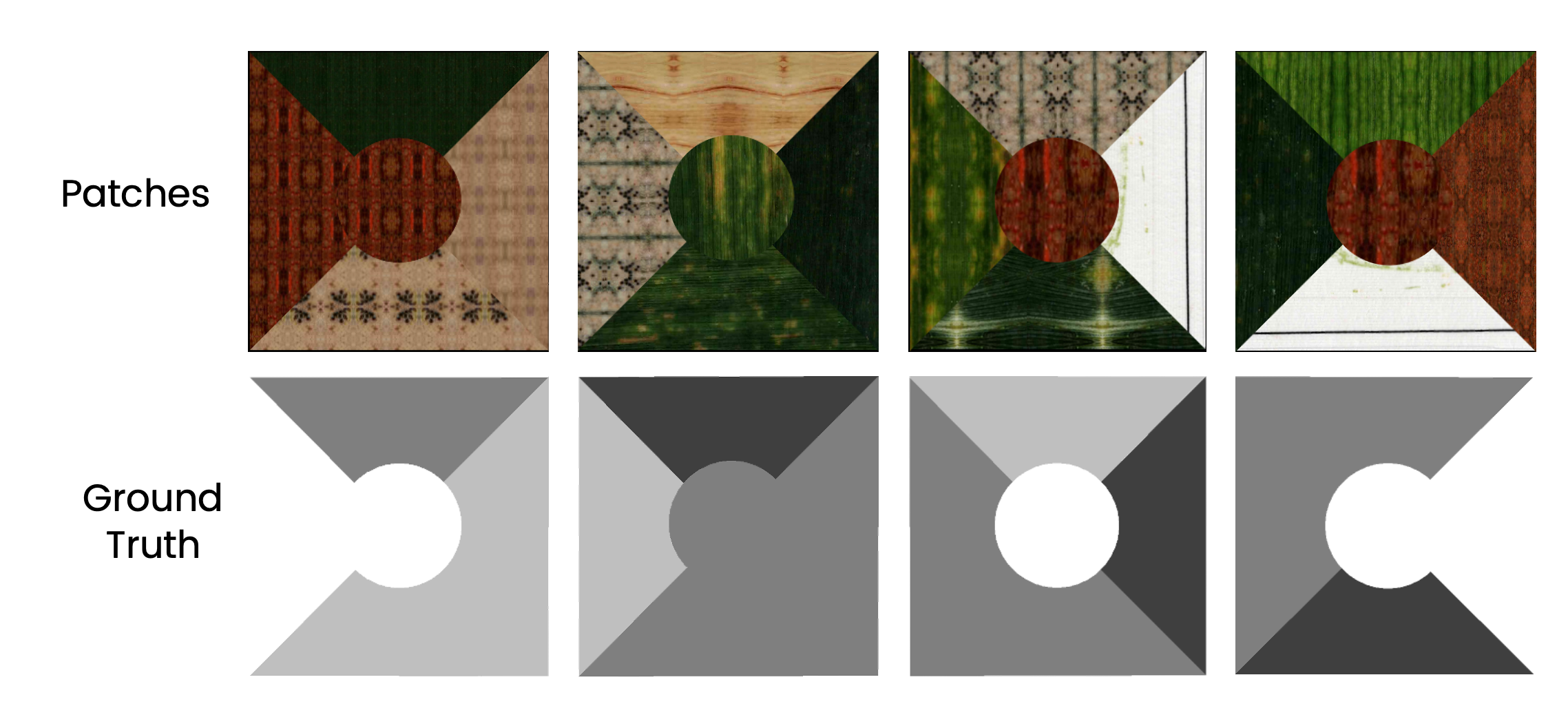
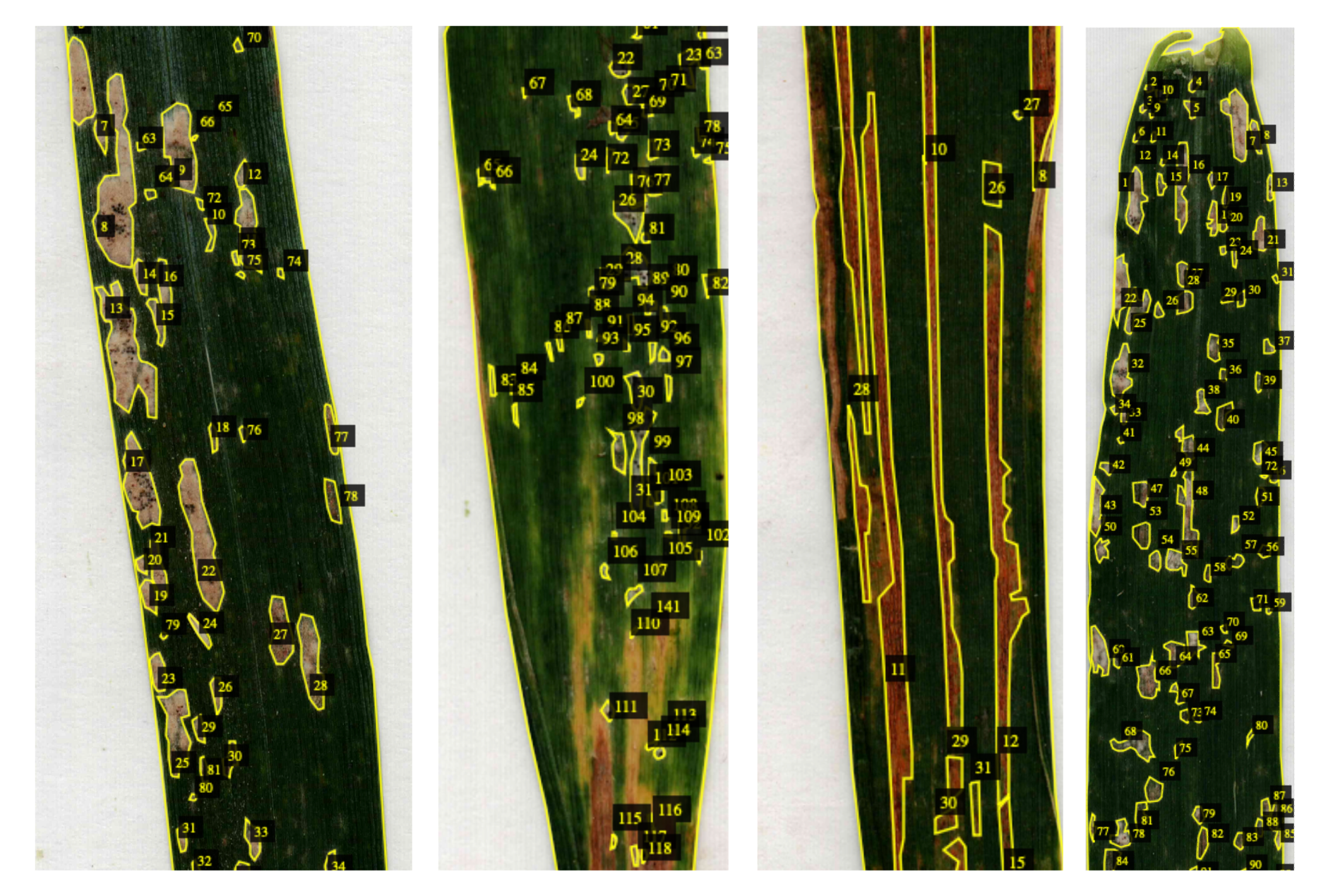
📈 Results
The model demonstrated good accuracy:
- On artificial patches, the model’s error in predicting infected areas was less than 1% compared to the ground truth.
- On real wheat leaves, the model’s prediction error was around 10%.
These results indicate that the model performs well in predicting the infected areas, with an acceptable margin of error according to the project’s requirements, making it a valuable tool for automating leaf analysis in agricultural research.
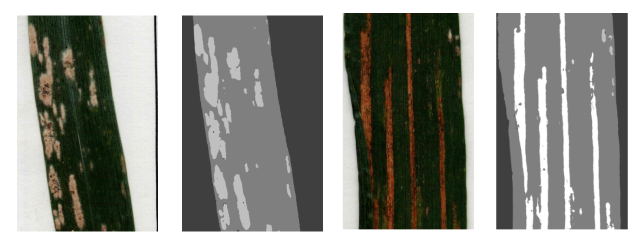
🚀 Real-world Application
Once the machine learning model is created, the pipeline is ready to be used in real-world applications. We have developed user-friendly tools to utilize this project:
- A GUI application
- A terminal command line tool
Both tools return the segmented leaves and a CSV file with the results, including the label of the leaf, the healthy area, the area of yellow rust, and the area of oidium.
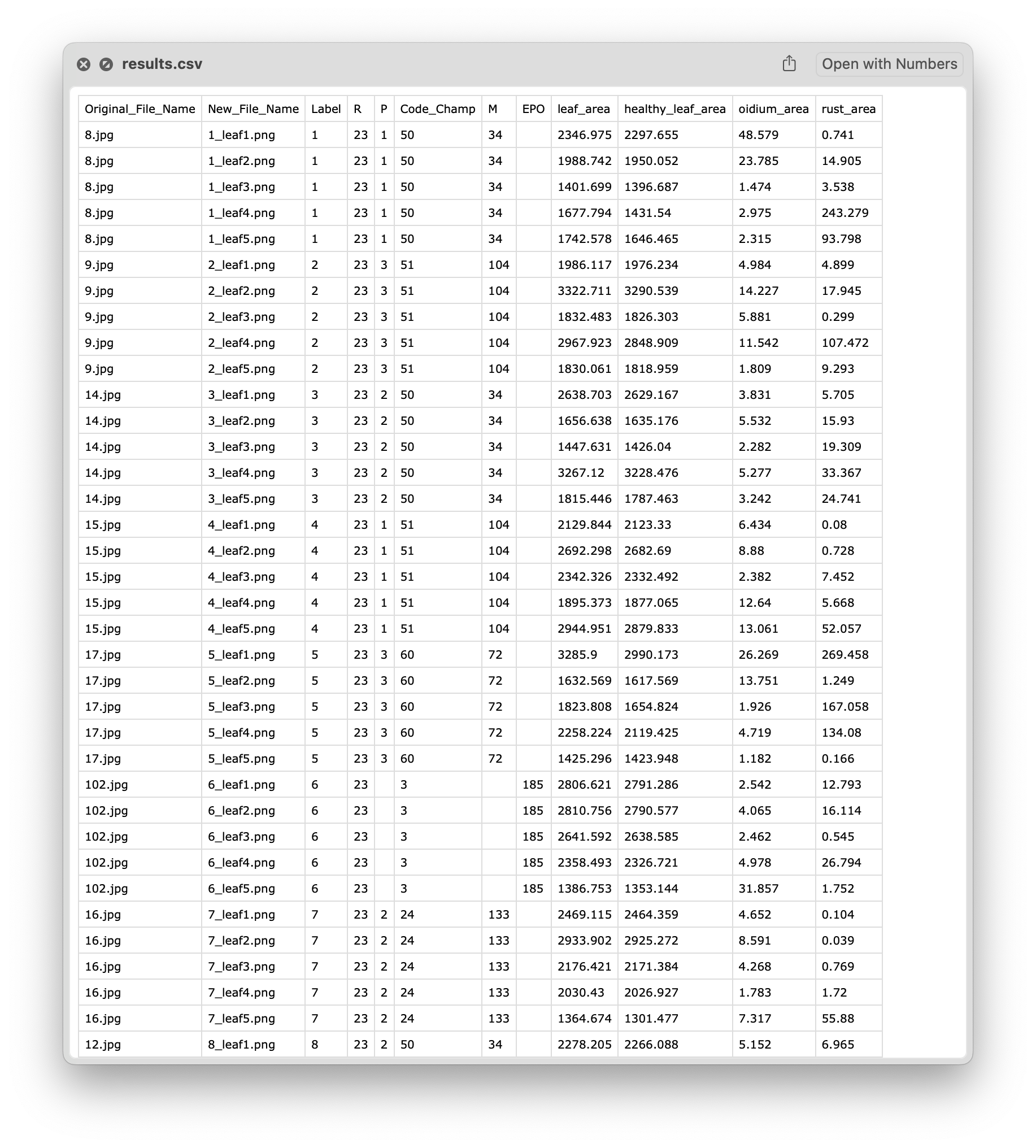
✅ Conclusion
Our machine learning model, developed in collaboration with Clément Plessis, Axel Prouvost, and Léna Connesson, has significantly improved the efficiency of wheat leaf analysis in the context of wheat variety mixture studies. By automating the process of identifying infected areas on wheat leaves, we have enabled researchers to analyze large datasets more quickly and accurately, facilitating the optimization of wheat variety mixtures for sustainable agriculture practices.
🚧 Challenges and Learnings
Challenges
Model Integration: Integrating Ilastik with Python workflows was initially challenging due to compatibility issues. Developing the EasIlastik package helped bridge this gap, but it required significant effort to ensure seamless operation.
Computational Constraints: Ensuring that the machine learning model could run efficiently on everyday laptops without powerful hardware or GPUs was a significant constraint. This required careful optimization of the model and preprocessing pipeline.
Learnings
Importance of Preprocessing: Effective preprocessing is crucial for the success of machine learning models, especially when dealing with real-world data that can be noisy and inconsistent.
Collaboration and Interdisciplinary Work: Collaborating with experts from different fields, such as genomics and computer science, was invaluable. Each team member brought unique insights that contributed to the project’s success.
Iterative Development: The project underscored the importance of iterative development and continuous testing. Regularly evaluating the model’s performance on both synthetic and real datasets helped identify and address issues early.
User-Friendly Tools: Developing tools that are easy to use and integrate into existing workflows is essential for adoption by researchers with varying levels of technical expertise. The EasIlastik package is an example of how simplifying complex processes can enhance usability and impact.
GitHub @titouanlegourrierec · Email titouanlegourrierec@icloud.com


